
In May 2023, the team of Tan Tao/Chen Yongchang/Ji Weizhi from Kunming University of Science and Technology, the team of Liu Bing from the Chinese People's Liberation Army (PLA) General Hospital and the team of Wu Jun from University of Texas, Southwestern Medical Center at Dallas collaborated to publish an article entitled Ex uteromonkey embryogenesis from blastocyst to early organogenesis in Cell (IF=66.850). This study developed an embedded 3D culture system, EMEUC, to achieve long time course culture of crab-eating macaque embryos up to 25 d.p.f. in vitro. Combined with morphological, histological and single-cell transcriptome sequencing analyses, we have systematically resolved the spectral specialization trajectories and molecular evolutionary patterns of early tissue and organogenesis, including protointestinal motility and hematopoiesis. ANNOROAD provided sequencing services for this study.
Research Background
There is a critical developmental milestone when quadrumana go through their third and fourth weeks of gestation, which culminates in the formation of the three major embryonic layers - ectoderm, mesoderm and endoderm - and the production of various organ prototypes. During this period, developmental failures and malformations can lead to early miscarriages and birth defects. Therefore, the exploration of embryonic regulatory mechanisms during this mysterious "black box" period is essential to improving our understanding of primate embryogenesis and to understanding the potential etiology of early developmental disorders.
The researchers developed an embedded 3D culture system, EMEUC, to optimize the pre-culture system of crab-eating macaque embryos and build a 3D culture environment with the help of extracellular matrix, finally realizing the 25-day in vitro long duration culture of crab-eating macaque embryos. Its morphology and developmental process are highly consistent with the in vivo embryo, which approximately corresponds to the CS8c stage in vivo.
Research Content
Single-cell Transcriptome Reveals Characteristics of EMEUC Embryos
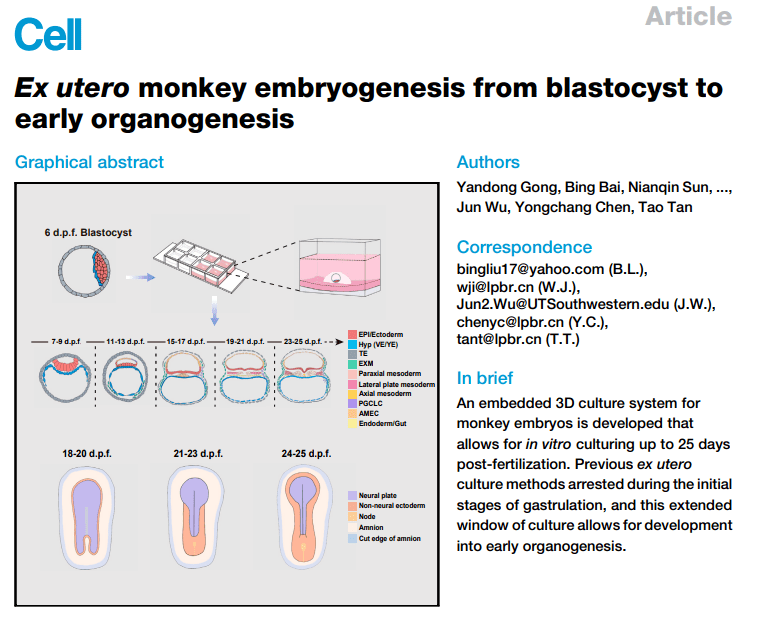
By sequencing the single-cell transcriptome of EMEUC-cultured crab-eating macaque embryos (hereafter referred to as EMEUC embryos) from 18 d.p.f. to 25 d.p.f., combined with the expression of significant Marker genes of known lineages, two taxa of hematopoietic cells and early intestinal cells were identified and the major 25 cell types were preliminarily annotated. All cells can be classified into five major categories: endoderm, ectoderm, trophectoderm, mesoderm and a mixed cell population including axial mesoderm (AM), EPI, neural plate, PGCLC, mesoendoderm (PS) and paraxial mesoderm (PM). Markers for progenitor cells are expressed in mixed cell populations, but not for fully differentiated cell types. In summary, the single-cell transcriptome sequencing profiles depict the major cell types in crab-eating macaques at the protointestinal embryo formation and early organogenesis stages.
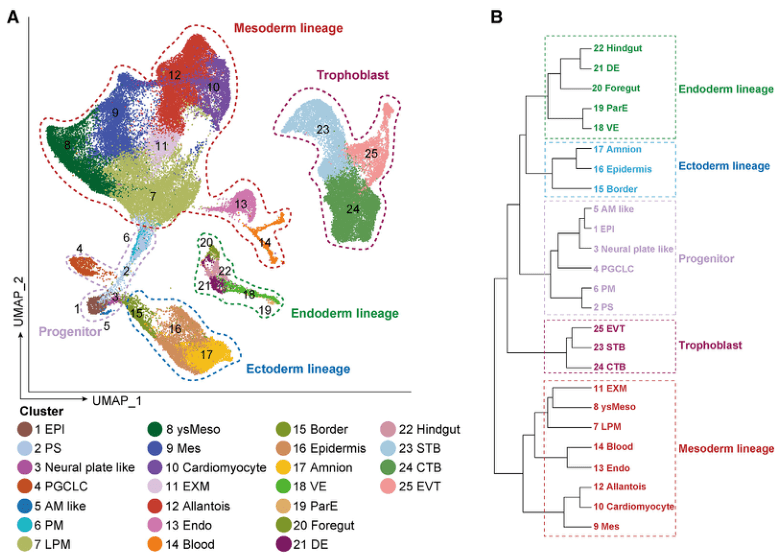
Figure Single-cell Transcriptome Analysis of EMEUC Embryos
Ectodermal Lineage Specialization in EMEUC Embryos - Early Neuroectodermal Development
Next, the investigators focused on the early neural differentiation of EMEUC embryos. Firstly, the Marker gene expression characteristics at different periods were further verified by immunofluorescence staining. Secondly, the proposed temporal analysis revealed the differentiation of EPI into NPPs and subsequently into NCPLCs and PPLCs, consistent with in vivo embryos. Thirdly, cell communication analysis demonstrated the activation of NODAL and WNT signaling pathways during neural lineage differentiation. Finally, SCENIC analysis identified transcription factor (TF) regulation and networks in NPP, NCPLCs, and PPLCs. In summary, EMEUC embryos recapitulate early neural ectodermal development, including transcriptional dynamics and spatial signaling interactions.
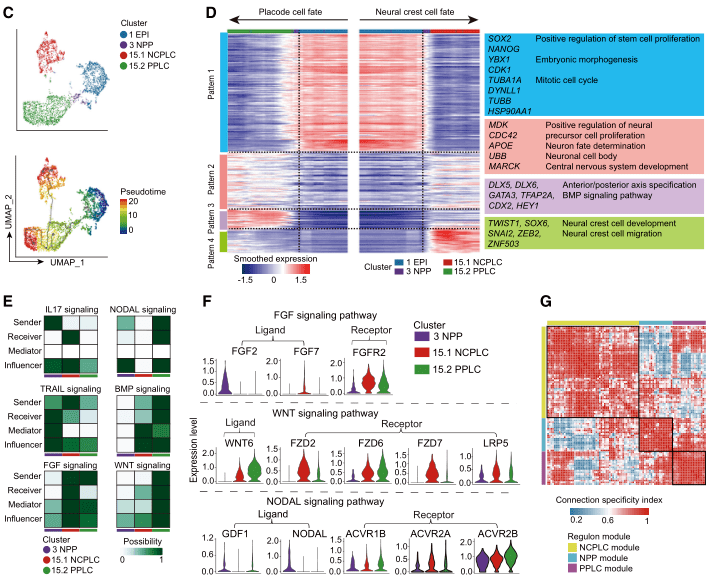
Figure Early Neurodevelopment of EMEUC Embryos
Mesodermal and Endodermal Lineage Specialization and Hematopoiesis of the Yolk Sac in EMEUC Embryos
EMEUC embryos similarly recapitulate the expression profile of the embryonic mesodermal lineage in vivo. The AM-like and PM cell populations are closely related to the EPI and PS cell populations, respectively, and have features that lack the expression of some mesodermal genes and are less differentiated. In contrast, LPM cells appear to have multiple mature cell types.
Hematopoietic lineage specialization in EMEUC embryos highly mimics the two waves of hematopoiesis in the yolk sac in vivo, recapitulating the early embryonic hematopoiesis. Two hematopoietic progenitor cells (cluster14.1 HPC1 and cluster14.3 HPC2) appear sequentially in the blood and have distinct transcriptome profiles. HPC1 resembles the first wave of hematopoietic progenitor cells by integration analysis with existing literature mouse and human early hematopoietic scRNA-seq datasets. HPC2 co-occurs with HEC and is closely related to the second wave of hematopoietic progenitor cells. EMEUC embryonic HEC are more similar to in vivo embryonic HEC than human pluripotent stem cell HEC.
In addition to the spectral specialization of mesoderm, EMEUC embryos also showed endodermal specialization, and the DE cells in the endodermal lineage (cluster21.1 DE1 and cluster21.2 DE2) resembled human proliferating DE cells. The foregut and hindgut subpopulations identified in EMEUC embryos were highly consistent with monkey embryos and human embryos in vivo. Further studies suggest that BMP and EGF signaling pathways are associated with foregut formation, while WNT and Hedgehog signaling pathways may play important roles in hindgut development.
Figure Mesodermal Lineage and Hematopoiesis of EMEUC Embryos
Development and X Chromosome Dosage Compensation Regulation of PGCLC Cells in EMEUC Embryos
Primordial germ cell-like cells (PGCLC) likewise undergo migration, differentiation and maturation in EMEUC embryos. Nine distinct subpopulations of PGCLC within EMEUC embryos are present almost throughout development (18-25 d.p.f.) and have distinct molecular characteristics. During the development, PGCLC cells began to migrate toward the yolk sac, and the expression of genes involved in oxidative phosphorylation was upregulated, while the expression of glycolysis-related genes was downregulated, suggesting a metabolic switch during early germ cell development in crab-eating macaques.
By comparing the X chromosome gene expression and X:A dosage balance in the internal and external lineages of embryos of different sexes at each period, this study also systematically revealed the X chromosome dosage compensation regulatory process in the early embryonic development of crab-eating macaques. The X chromosome gene expression level is gradually up-regulated in female PGCLC, and there is an X chromosome reactivation process. XIST, which is involved in X chromosome inactivation, is rarely or not expressed in male early PGCLCs and elevated in late stages, while the expression level is relatively constant in female PGCLCs.
Research Findings
This study developed a 3D culture system capable of supporting the development of crab-eating macaque embryos in vitro to early organogenesis, successfully extended the embryonic development of crab-eating macaques in vitro to the early organogenesis stage (~25 d.p.f.), and systematically investigated the important cell composition and spectral differentiation trajectory of non-human primates from protointestinal embryo formation to early organogenesis, which will contribute to the understanding of human embryonic developmental malformations and etiology of early pregnancy miscarriage.