Brief
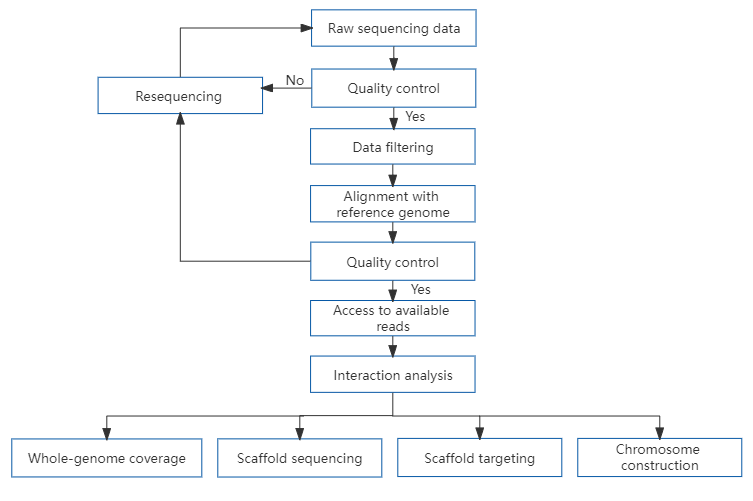
Hi-C technology is derived from chromosome conformation capture technology, adopts high-throughput sequencing technology, and combines bioinformatics analysis methods to study the spatial relationship of the entire chromatin DNA in the whole genome, and obtain high-resolution chromatin three-dimensional structure information. Based on the fact that the interaction intensity between chromatin fragments in Hi-C data decreases with distance, Hi-C can be used for genome assembly, assembling chaotic gene sequences to the chromosomal level.